Bipartite monolayer network example
Data set
A weighted bipartite network describing a plant-flower visitor
interaction web (25 plant species and 79 flower visitor species) in the
vicinity of Bristol, U.K. To distinguish between the two node sets we
number the pollinator species from 1-79 and the plants from 80-104. Data
can be obtained using data(memmott1999) in package
bipartite.
Input
A link
list with columns from, to and
weight. Because this is a bipartite network the
from column can only contain nodes 1-79 and the
to column nodes 84-104.
R Code
# Import data
library(infomapecology)
library(bipartite)
data(memmott1999)
network_object <- create_monolayer_network(memmott1999, bipartite = T, directed = F, group_names = c('A','P'))## [1] "Input: a bipartite matrix"infomap_object <- run_infomap_monolayer(network_object, infomap_executable='Infomap',
flow_model = 'undirected',
silent=T, trials=20, two_level=T, seed=123)## [1] "Creating a link list..."
## running: ./Infomap infomap.txt . --tree --seed 123 -N 20 -f undirected --silent --two-level
## [1] "Removing auxilary files..."# Plot the matrix (plotting function in beta)
plot_modular_matrix(infomap_object)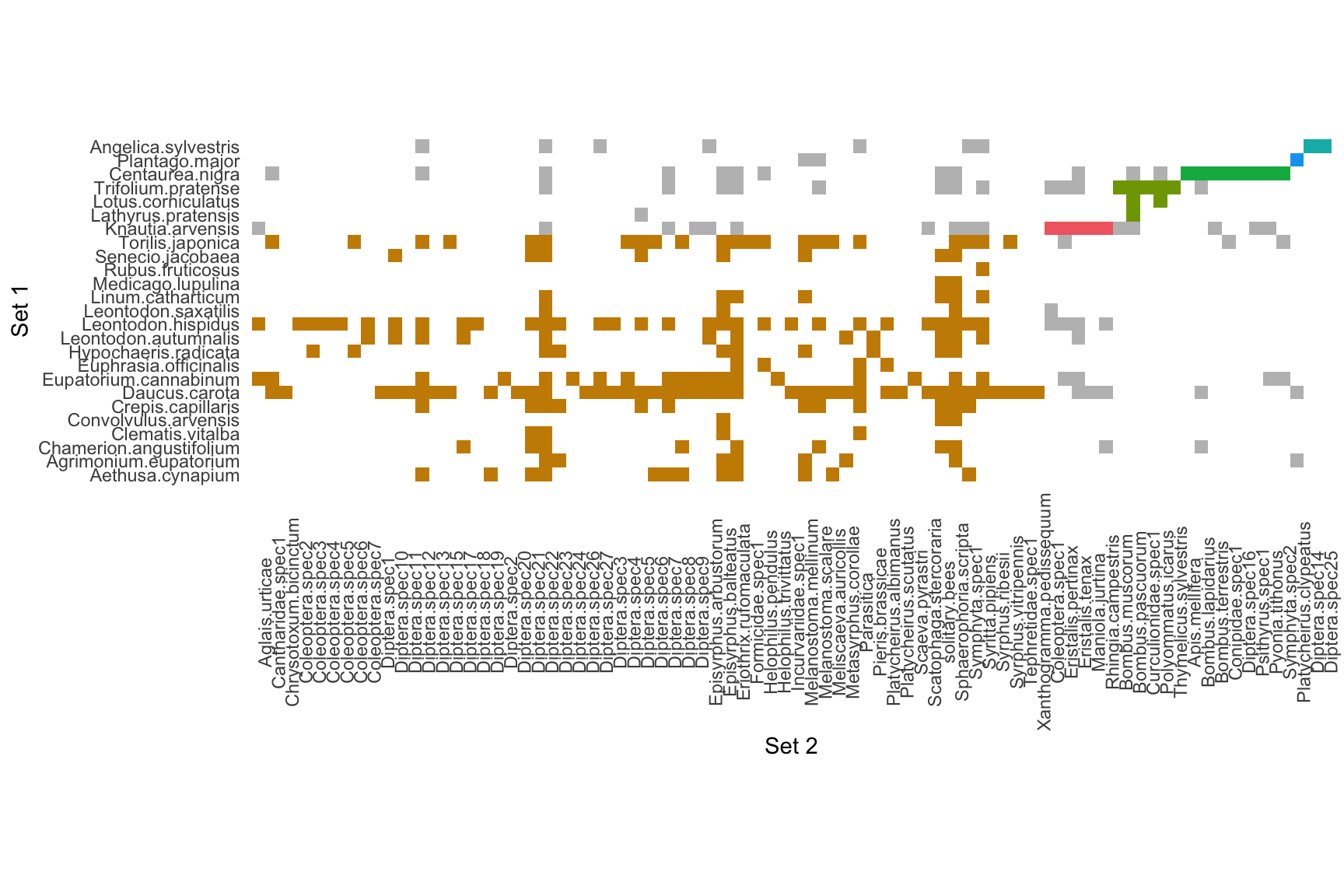
Infomap
Under the hood, the function run_infomap_monolayer
runs:
./Infomap infomap.txt . --tree -i link-list --seed 123 -N 20 -f undirected -2With this command, Infomap detects modules that contain both plants
and pollinators. * -f undirected indicates flow on an
undirected network. * -2 indicates a two-level solution,
with no hierarchical modules.
Output
A tree
file is produced by Infomap, but is parsed by
run_infomap_monolayer from infomapecology (in R:
?run_infomap_monolayer).